
Chapter 14 : Pandas User Defined Function¶
Chapter Learning Objectives¶
Various data operations using Pandas User Defined Function.
Chapter Outline¶
import pyspark
from pyspark.sql import SparkSession
spark = SparkSession \
.builder \
.appName("Python Spark SQL basic example") \
.config("spark.some.config.option", "some-value") \
.getOrCreate()
from IPython.display import display_html
import pandas as pd
import numpy as np
def display_side_by_side(*args):
html_str=''
for df in args:
html_str+=df.to_html(index=False)
html_str+= "\xa0\xa0\xa0"*10
display_html(html_str.replace('table','table style="display:inline"'),raw=True)
space = "\xa0" * 10
import panel as pn
css = """
div.special_table + table, th, td {
border: 3px solid orange;
}
"""
pn.extension(raw_css=[css])
Pandas User Defined Function¶
A pandas user-defined function (UDF)—also known as vectorized UDF—is a user-defined function that uses Apache Arrow to transfer data and pandas to work with the data. pandas UDFs allow vectorized operations that can increase performance up to 100x compared to row-at-a-time Python UDFs.
Pandas is well known to data scientists and has seamless integrations with many Python libraries and packages such as NumPy, statsmodel, and scikit-learn, and Pandas UDFs allow data scientists not only to scale out their workloads, but also to leverage the Pandas APIs in Apache Spark.
The user-defined functions are executed by:
Apache Arrow, to exchange data directly between JVM and Python driver/executors with near-zero (de)serialization cost. Pandas inside the function, to work with Pandas instances and APIs. The Pandas UDFs work with Pandas APIs inside the function and Apache Arrow for exchanging data. It allows vectorized operations that can increase performance up to 100x, compared to row-at-a-time Python UDF
Python Type Hints
Python type hints were officially introduced in PEP 484 with Python 3.5. Type hinting is an official way to statically indicate the type of a value in Python. See the example below.
def greeting(name: str) -> str: return ‘Hello ‘ + name The name: strindicates the name argument is of str type and the -> syntax indicates the greeting() function returns a string.
Python type hints bring two significant benefits to the PySpark and Pandas UDF context.
It gives a clear definition of what the function is supposed to do, making it easier for users to understand the code. For example, unless it is documented, users cannot know if greeting can take None or not if there is no type hint. It can avoid the need to document such subtle cases with a bunch of test cases and/or for users to test and figure out by themselves. It can make it easier to perform static analysis. IDEs such as PyCharm and Visual Studio Code can leverage type annotations to provide code completion, show errors, and support better go-to-definition functionality.
There are currently four supported cases of the Python type hints in Pandas UDFs:
Series to Series
Iterator of Series to Iterator of Series
Iterator of Multiple Series to Iterator of Series
Series to Scalar (a single value)
Before we do a deep dive into each case, let’s look at three key points about working with the new Pandas UDFs.
Although Python type hints are optional in the Python world in general, you must specify Python type hints for the input and output in order to use the new Pandas UDFs.
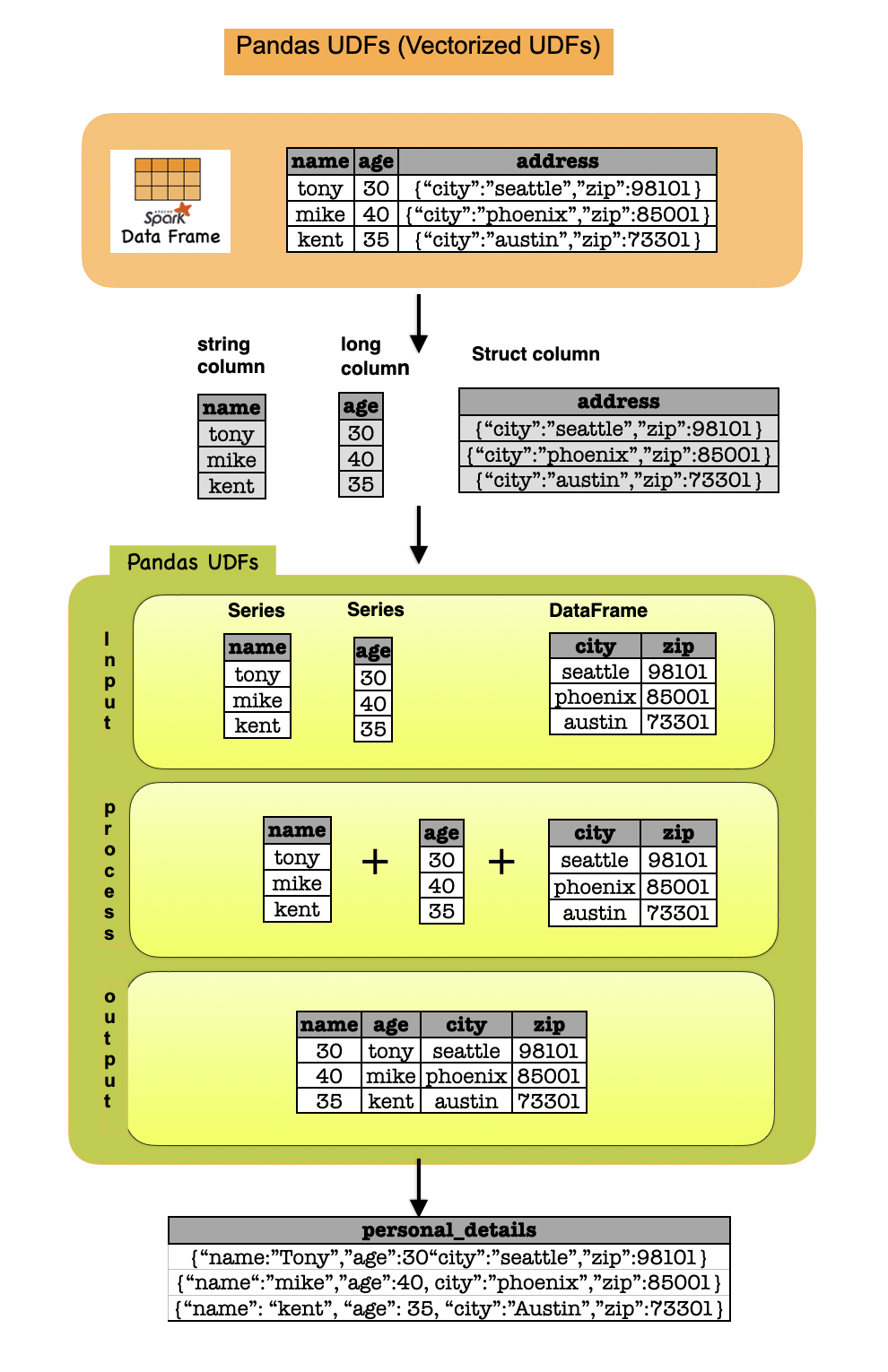
The type hint should use pandas.Series in all cases. However, there is one variant in which pandas.DataFrame should be used for its input or output type hint instead: when the input or output column is of StructType.Take a look at the example below:
import numpy as np
import pandas as pd
# Enable Arrow-based columnar data transfers
spark.conf.set("spark.sql.execution.arrow.pyspark.enabled", "true")
# Generate a Pandas DataFrame
pdf = pd.DataFrame(np.random.rand(100, 3))
# Create a Spark DataFrame from a Pandas DataFrame using Arrow
df = spark.createDataFrame(pdf)
# Convert the Spark DataFrame back to a Pandas DataFrame using Arrow
result_pdf = df.select("*").toPandas()
print("Pandas DataFrame result statistics:\n%s\n" % str(result_pdf.describe()))
Pandas DataFrame result statistics:
0 1 2
count 100.000000 100.000000 100.000000
mean 0.475495 0.497599 0.518556
std 0.294840 0.277754 0.298373
min 0.011082 0.017651 0.012683
25% 0.222519 0.256175 0.250417
50% 0.444332 0.503060 0.545583
75% 0.735770 0.736086 0.778632
max 0.989444 0.999400 0.984667
Using the above optimizations with Arrow will produce the same results as when Arrow is not enabled.
Note that even with Arrow, toPandas() results in the collection of all records in the DataFrame to the driver program and should be done on a small subset of the data.
Not all Spark data types are currently supported and an error can be raised if a column has an unsupported type, see Supported SQL Types. If an error occurs during createDataFrame(), Spark will fall back to create the DataFrame without Arrow.
Supported SQL Types
Currently, all Spark SQL data types are supported by Arrow-based conversion except MapType, ArrayType of TimestampType, and nested StructType.
1a. Pandas UDFs (Vectorized UDFs)¶

Pandas UDFs are user defined functions that are executed by Spark using Arrow to transfer data to perform vectorized operations.
A Pandas UDF is defined using the pandas_udf as a decorator.
Pandas UDFs used to be defined with Python type hints.
Note that the type hint should use pandas.Series in all cases but there is one variant that pandas.DataFrame should be used for its input or output type hint instead when the input or output column is of StructType.
The following example shows a Pandas UDF which takes long column, string column and struct column, and outputs a struct column. It requires the function to specify the type hints of pandas.Series and pandas.DataFrame as below:
Lets first understand the syntax
Syntax
pyspark.sql.functions.pandas_udf(f=None, returnType=None, functionType=None)
Creates a pandas user defined function (a.k.a. vectorized user defined function).
Parameters
f – user-defined function. A python function if used as a standalone function
returnType – the return type of the user-defined function. The value can be either a pyspark.sql.types.DataType object or a DDL-formatted type string.
functionType – an enum value in pyspark.sql.functions.PandasUDFType. Default: SCALAR. ‘’’
import pandas as pd
from pyspark.sql.functions import pandas_udf
@pandas_udf("city string, col2 long")
def func(s1: pd.Series, s2: pd.Series, s3: pd.DataFrame) -> pd.DataFrame:
s3['col2'] = s1 + s2.str.len()
return s3
# Create a Spark DataFrame that has three columns including a sturct column.
df = spark.createDataFrame(
[[1, "tony", ("seattle",)]],
"id long, name string, city_struct struct<city:string>")
df.printSchema()
df.show()
df_pandas = df.select(func("id", "name", "city_struct").alias("pandas_udf"))
df_pandas.printSchema()
df_pandas.toPandas()#show()
root
|-- id: long (nullable = true)
|-- name: string (nullable = true)
|-- city_struct: struct (nullable = true)
| |-- city: string (nullable = true)
+---+----+-----------+
| id|name|city_struct|
+---+----+-----------+
| 1|tony| [seattle]|
+---+----+-----------+
root
|-- pandas_udf: struct (nullable = true)
| |-- city: string (nullable = true)
| |-- col2: long (nullable = true)
| pandas_udf | |
|---|---|
| 0 | {'city': 'seattle', 'col2': 5} |
Summary:
print("Input ", "Output")
display_side_by_side(df.toPandas(),df_pandas.toPandas())
Input Output
| id | name | city_struct |
|---|---|---|
| 1 | tony | {'city': 'seattle'} |
| pandas_udf |
|---|
| {'city': 'seattle', 'col2': 5} |
display_html(df.printSchema())
display_html(df.printSchema())
root
|-- id: long (nullable = true)
|-- name: string (nullable = true)
|-- city_struct: struct (nullable = true)
| |-- city: string (nullable = true)
root
|-- id: long (nullable = true)
|-- name: string (nullable = true)
|-- city_struct: struct (nullable = true)
| |-- city: string (nullable = true)
var1 = df._jdf.schema().treeString()
print(var1,end ='');
print(var1),
root
|-- id: long (nullable = true)
|-- name: string (nullable = true)
|-- city_struct: struct (nullable = true)
| |-- city: string (nullable = true)
root
|-- id: long (nullable = true)
|-- name: string (nullable = true)
|-- city_struct: struct (nullable = true)
| |-- city: string (nullable = true)
(None,)
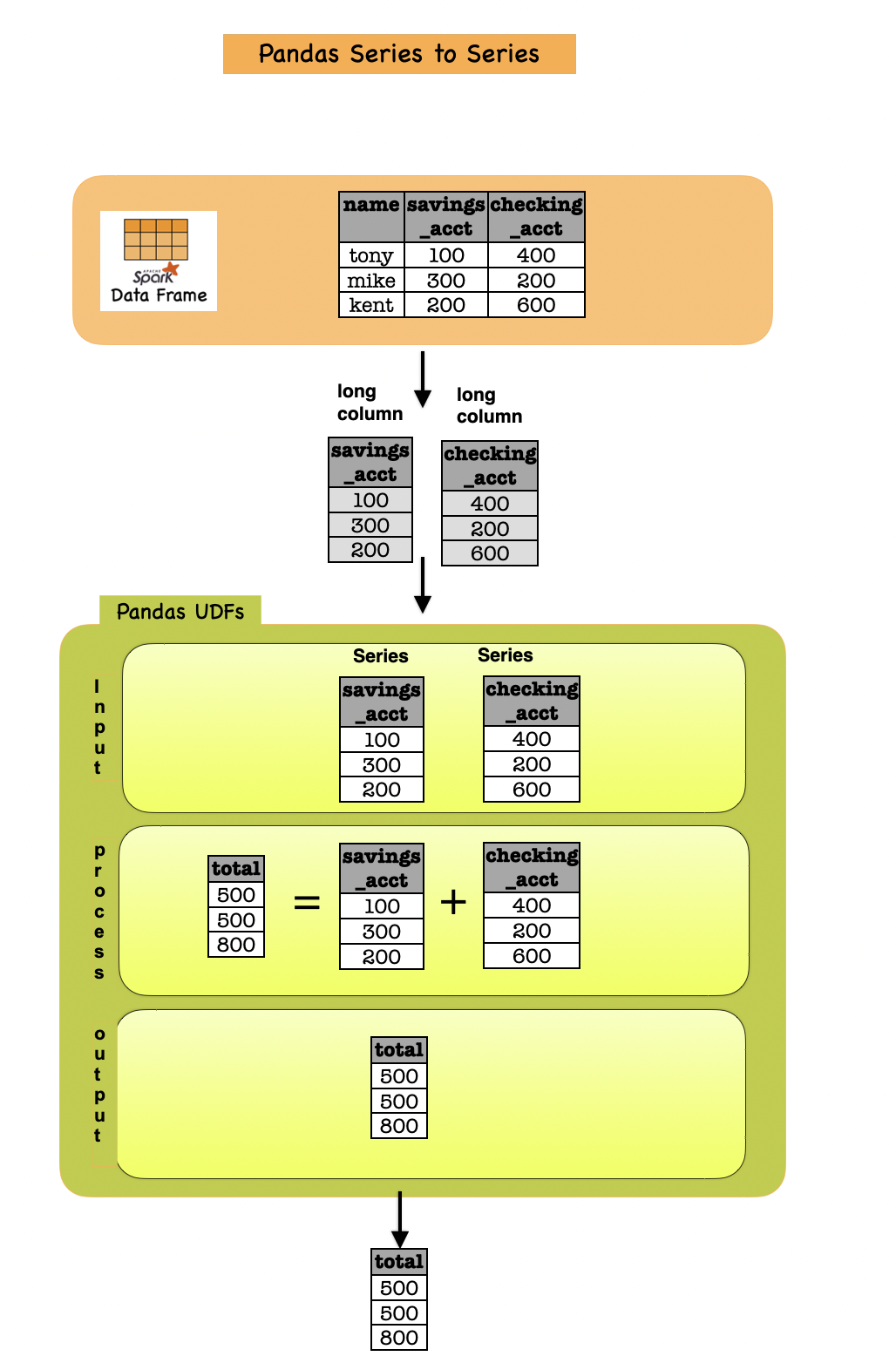
1b. Pandas UDF: Series to Series¶
import pandas as pd
from pyspark.sql.functions import col, pandas_udf
from pyspark.sql.types import LongType
# Declare the function and create the UDF
def multiply_func(a: pd.Series, b: pd.Series) -> pd.Series:
return a * b
multiply = pandas_udf(multiply_func, returnType=LongType())
# The function for a pandas_udf should be able to execute with local Pandas data
x = pd.Series([1, 2, 3])
print(multiply_func(x, x))
# 0 1
# 1 4
# 2 9
# dtype: int64
# Create a Spark DataFrame, 'spark' is an existing SparkSession
df = spark.createDataFrame(pd.DataFrame(x, columns=["x"]))
# Execute function as a Spark vectorized UDF
df.select(multiply(col("x"), col("x"))).show()
# +-------------------+
# |multiply_func(x, x)|
# +-------------------+
# | 1|
# | 4|
# | 9|
# +-------------------+
0 1
1 4
2 9
dtype: int64
+-------------------+
|multiply_func(x, x)|
+-------------------+
| 1|
| 4|
| 9|
+-------------------+
1c. Iterator of Series to Iterator of Series¶
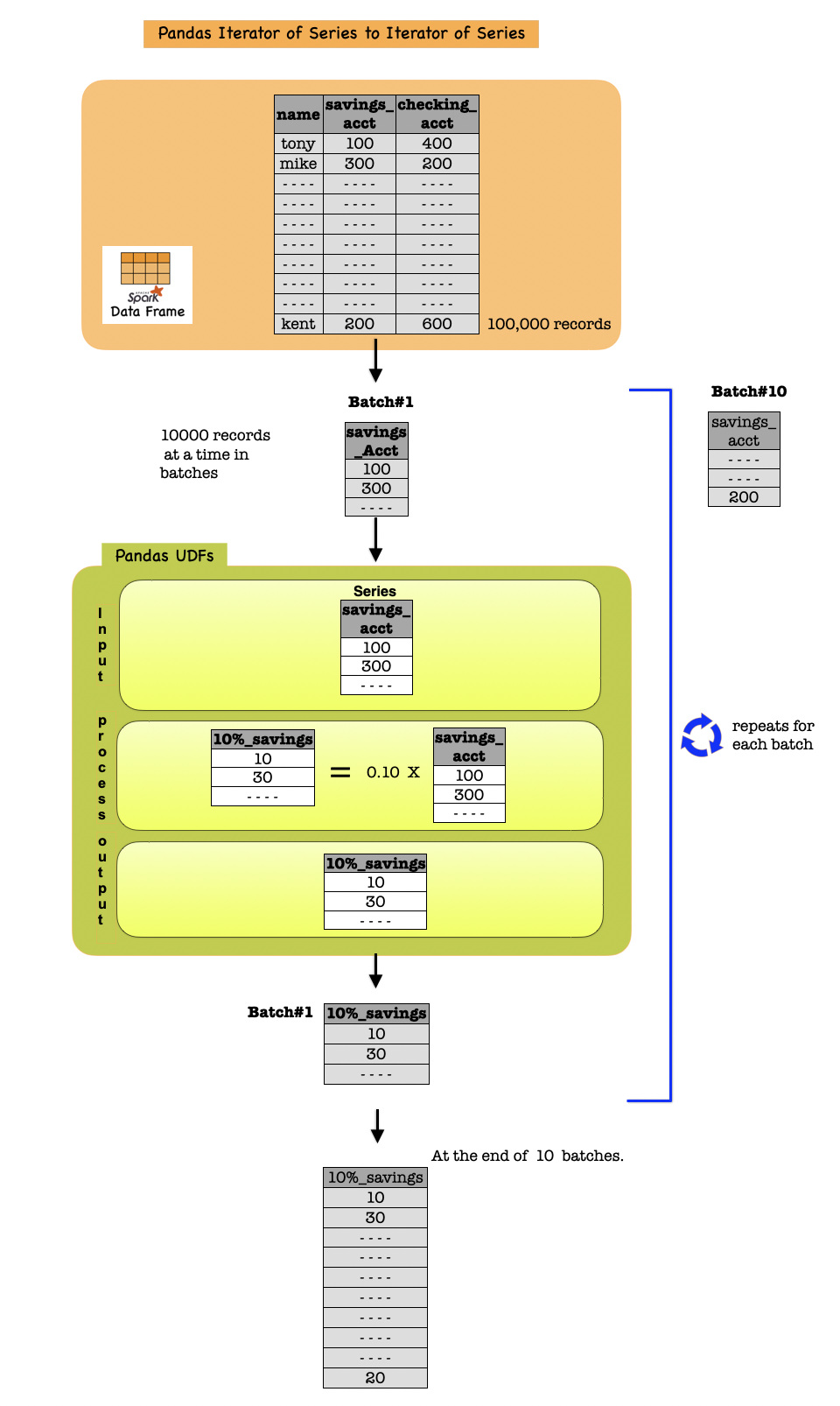
An iterator UDF is the same as a scalar pandas UDF except:
The Python function
Takes an iterator of batches instead of a single input batch as input.
Returns an iterator of output batches instead of a single output batch.
The length of the entire output in the iterator should be the same as the length of the entire input.
The wrapped pandas UDF takes a single Spark column as an input.
You should specify the Python type hint as Iterator[pandas.Series] -> Iterator[pandas.Series].
This pandas UDF is useful when the UDF execution requires initializing some state, for example, loading a machine learning model file to apply inference to every input batch.
The following example shows how to create a pandas UDF with iterator support.
from typing import Iterator
import pandas as pd
from pyspark.sql.functions import pandas_udf
pdf = pd.DataFrame([1, 2, 3], columns=["x"])
df = spark.createDataFrame(pdf)
df.show()
var_bc = spark.sparkContext.broadcast(100)
def calculate_complex(var1,var2):
return var1+var2+var1*var2
# Declare the function and create the UDF
@pandas_udf("long")
def plus_one(iterator: Iterator[pd.Series]) -> Iterator[pd.Series]:
var = var_bc.value
for x in iterator:
yield calculate_complex(x , var)
df_out = df.select(plus_one("x"))
df_out.show()
+---+
| x|
+---+
| 1|
| 2|
| 3|
+---+
+-----------+
|plus_one(x)|
+-----------+
| 201|
| 302|
| 403|
+-----------+
print("Input ", "Output")
display_side_by_side(df.toPandas(),df_out.toPandas())
Input Output
| x |
|---|
| 1 |
| 2 |
| 3 |
| plus_one(x) |
|---|
| 201 |
| 302 |
| 403 |
1d. Iterator of Multiple Series to Iterator of Series¶
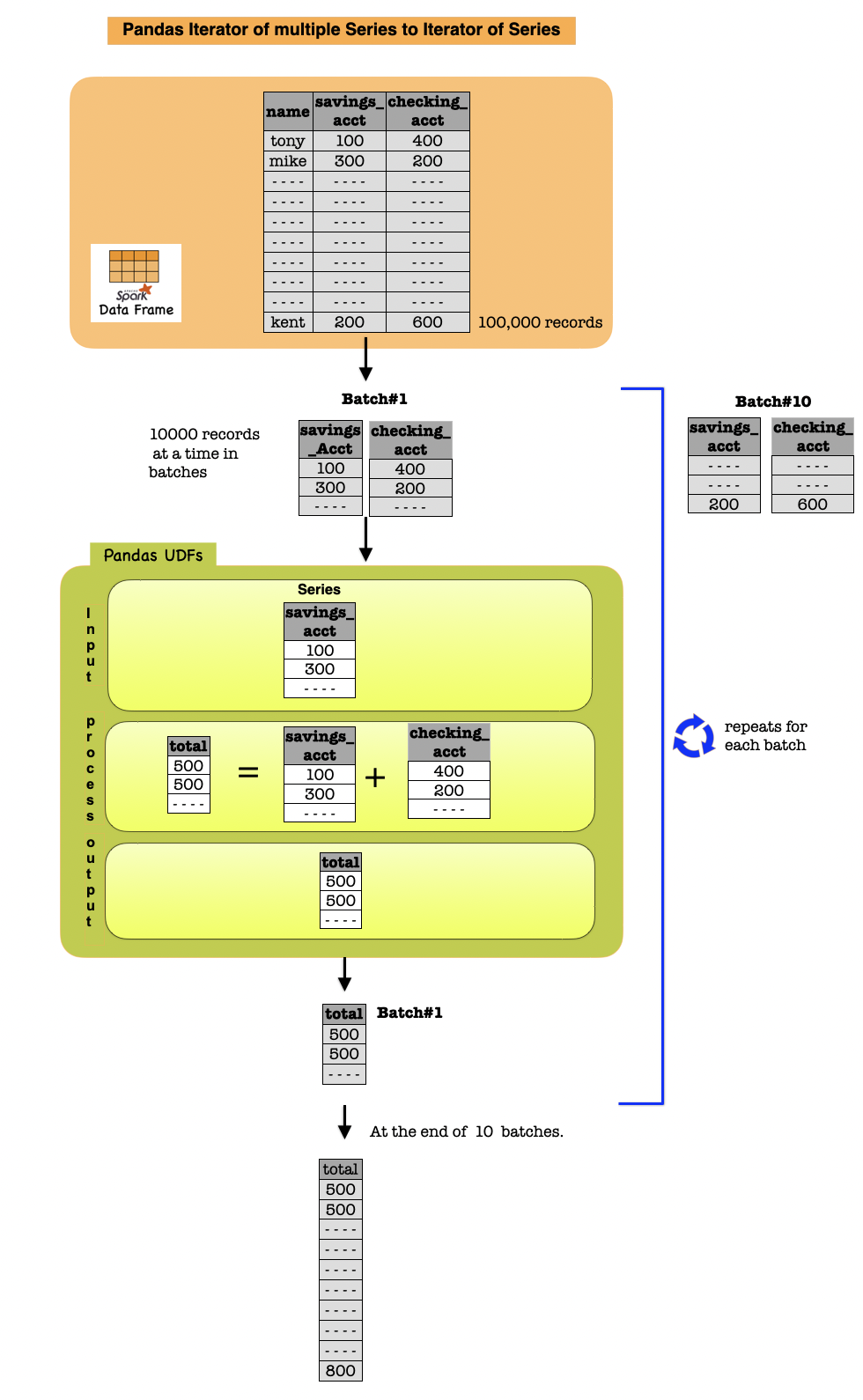
An Iterator of multiple Series to Iterator of Series UDF has similar characteristics and restrictions as Iterator of Series to Iterator of Series UDF.
The specified function takes an iterator of batches and outputs an iterator of batches.
It is also useful when the UDF execution requires initializing some state.
The differences are:
The underlying Python function takes an iterator of a tuple of pandas Series.
The wrapped pandas UDF takes multiple Spark columns as an input.
Input: Spark data frame with map column
from typing import Iterator, Tuple
import pandas as pd
from pyspark.sql.functions import pandas_udf
pdf = pd.DataFrame([(1,2,),(3,4,),(5,6)], columns=["weight","height"])
df = spark.createDataFrame(pdf)
var_bc = spark.sparkContext.broadcast(5)
def calculate_complex_mul(var1,var2,var3):
return var1+var2+var3
# # Declare the function and create the UDF
@pandas_udf("long")
def run_ml_model(iterator: Iterator[Tuple[pd.Series, pd.Series]]) -> Iterator[pd.Series]:
var = var_bc.value
for a, b in iterator:
yield calculate_complex_mul(a,b,var)
df_out_mul = df.select(run_ml_model("weight", "height"))
print("Input ", "Output")
display_side_by_side(df.toPandas(),df_out_mul.toPandas())
Input Output
| weight | height |
|---|---|
| 1 | 2 |
| 3 | 4 |
| 5 | 6 |
| run_ml_model(weight, height) |
|---|
| 8 |
| 12 |
| 16 |
1e. Series to Scalar¶
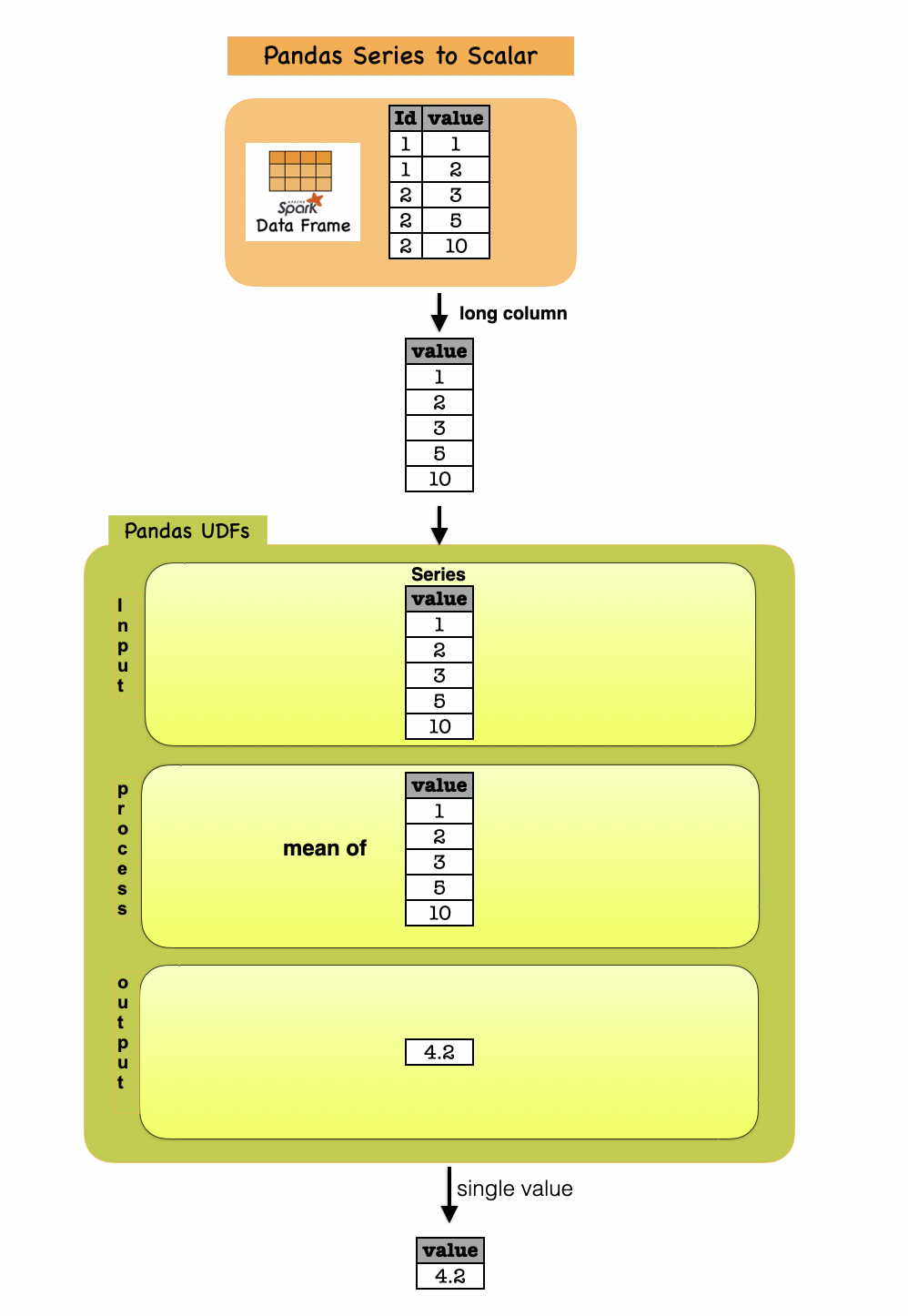
import pandas as pd
from pyspark.sql.functions import pandas_udf
from pyspark.sql import Window
df = spark.createDataFrame(
[(1, 1.0), (1, 2.0), (2, 3.0), (2, 5.0), (2, 10.0)],
("id", "v"))
# Declare the function and create the UDF
@pandas_udf("double")
def mean_udf(v: pd.Series) -> float:
return v.mean()
df_sca = df.select(mean_udf(df['v']))
print("Input ", "Output")
display_side_by_side(df.toPandas(),df_sca.toPandas())
Input Output
| id | v |
|---|---|
| 1 | 1.0 |
| 1 | 2.0 |
| 2 | 3.0 |
| 2 | 5.0 |
| 2 | 10.0 |
| mean_udf(v) |
|---|
| 4.2 |
df.groupby("id").agg(mean_udf(df['v'])).show()
w = Window \
.partitionBy('id') \
.rowsBetween(Window.unboundedPreceding, Window.unboundedFollowing)
df.withColumn('mean_v', mean_udf(df['v']).over(w)).show()
+---+-----------+
| id|mean_udf(v)|
+---+-----------+
| 1| 1.5|
| 2| 6.0|
+---+-----------+
+---+----+------+
| id| v|mean_v|
+---+----+------+
| 1| 1.0| 1.5|
| 1| 2.0| 1.5|
| 2| 3.0| 6.0|
| 2| 5.0| 6.0|
| 2|10.0| 6.0|
+---+----+------+
1f. Grouped Map¶
Grouped map operations with Pandas instances are supported by DataFrame.groupby().applyInPandas() which requires a Python function that takes a pandas.DataFrame and return another pandas.DataFrame. It maps each group to each pandas.DataFrame in the Python function.
This API implements the “split-apply-combine” pattern which consists of three steps:
Split the data into groups by using DataFrame.groupBy.
Apply a function on each group. The input and output of the function are both pandas.DataFrame. The input data - contains all the rows and columns for each group.
Combine the results into a new PySpark DataFrame.
To use groupBy().applyInPandas(), the user needs to define the following:
A Python function that defines the computation for each group. A StructType object or a string that defines the schema of the output PySpark DataFrame. The column labels of the returned pandas.DataFrame must either match the field names in the defined output schema if specified as strings, or match the field data types by position if not strings, e.g. integer indices. See pandas.DataFrame on how to label columns when constructing a pandas.DataFrame.
Note that all data for a group will be loaded into memory before the function is applied. This can lead to out of memory exceptions, especially if the group sizes are skewed. The configuration for maxRecordsPerBatch is not applied on groups and it is up to the user to ensure that the grouped data will fit into the available memory.
The following example shows how to use groupby().applyInPandas() to subtract the mean from each value in the group.
Setting Arrow Batch Size
Data partitions in Spark are converted into Arrow record batches, which can temporarily lead to high memory usage in the JVM. To avoid possible out of memory exceptions, the size of the Arrow record batches can be adjusted by setting the conf “spark.sql.execution.arrow.maxRecordsPerBatch” to an integer that will determine the maximum number of rows for each batch. The default value is 10,000 records per batch. If the number of columns is large, the value should be adjusted accordingly. Using this limit, each data partition will be made into 1 or more record batches for processing.
Syntax
pyspark.sql.GroupedData.applyInPandas(func, schema)
Maps each group of the current DataFrame using a pandas udf and returns the result as a DataFrame.
The function should take a pandas.DataFrame and return another pandas.DataFrame. For each group, all columns are passed together as a pandas.DataFrame to the user-function and the returned pandas.DataFrame are combined as a DataFrame.
The schema should be a StructType describing the schema of the returned pandas.DataFrame. The column labels of the returned pandas.DataFrame must either match the field names in the defined schema if specified as strings, or match the field data types by position if not strings, e.g. integer indices. The length of the returned pandas.DataFrame can be arbitrary.
Parameters
func – a Python native function that takes a pandas.DataFrame, and outputs a pandas.DataFrame.
schema – the return type of the func in PySpark. The value can be either a pyspark.sql.types.DataType object or a DDL-formatted type string.
‘’’
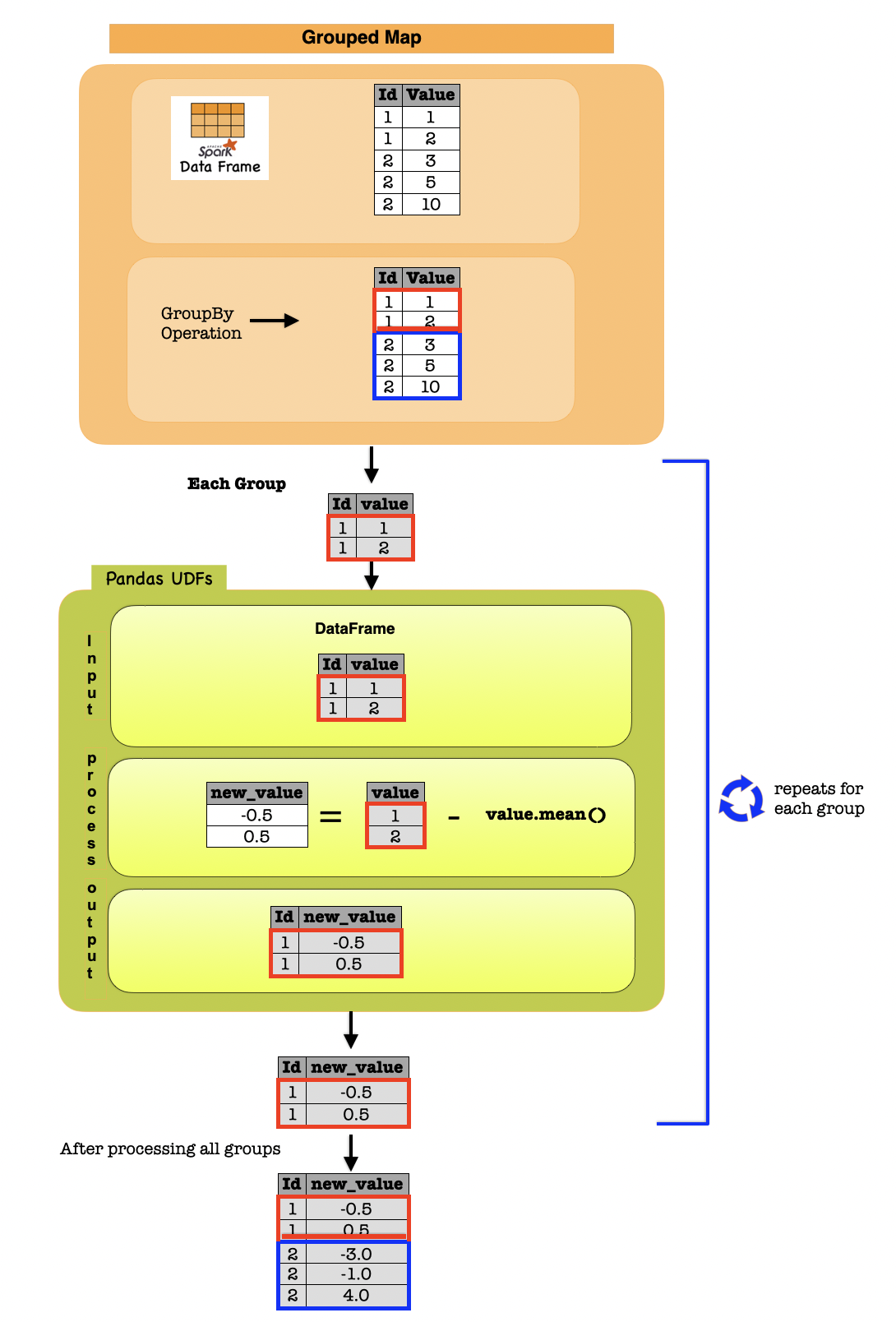
df = spark.createDataFrame(
[(1, 1.0), (1, 2.0), (2, 3.0), (2, 5.0), (2, 10.0)],
("id", "value"))
def subtract_mean(pdf):
# pdf is a pandas.DataFrame
value = pdf.value
return pdf.assign(value=value - value.mean())
df_group_pandas = df.groupby("id").applyInPandas(subtract_mean, schema="id long, value double")
print("Input ", "Output")
display_side_by_side(df.toPandas(),df_group_pandas.toPandas())
Input Output
| id | value |
|---|---|
| 1 | 1.0 |
| 1 | 2.0 |
| 2 | 3.0 |
| 2 | 5.0 |
| 2 | 10.0 |
| id | value |
|---|---|
| 1 | -0.5 |
| 1 | 0.5 |
| 2 | -3.0 |
| 2 | -1.0 |
| 2 | 4.0 |
1g. Map¶
Map operations with Pandas instances are supported by DataFrame.mapInPandas() which maps an iterator of pandas.DataFrames to another iterator of pandas.DataFrames that represents the current PySpark DataFrame and returns the result as a PySpark DataFrame.
The functions takes and outputs an iterator of pandas.DataFrame.
It can return the output of arbitrary length in contrast to some Pandas UDFs although internally it works similarly with Series to Series Pandas UDF.
The following example shows how to use mapInPandas():
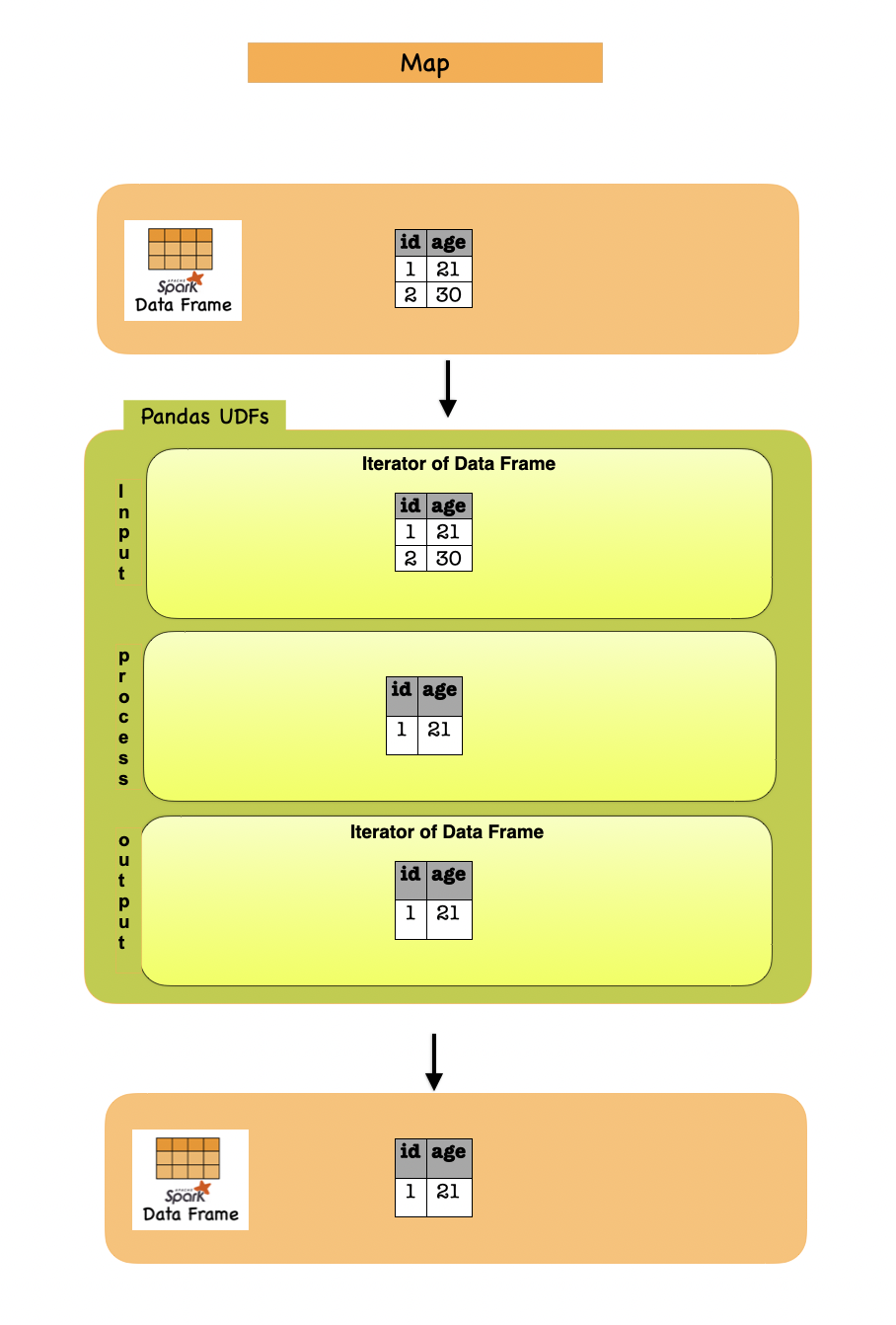
Lets first understand the syntax
Syntax
pyspark.sql.DataFrame.mapInPandas(func, schema)
Maps an iterator of batches in the current DataFrame using a Python native function that takes and outputs a pandas DataFrame, and returns the result as a DataFrame.
The function should take an iterator of pandas.DataFrames and return another iterator of pandas.DataFrames. All columns are passed together as an iterator of pandas.DataFrames to the function and the returned iterator of pandas.DataFrames are combined as a DataFrame. Each pandas.DataFrame size can be controlled by spark.sql.execution.arrow.maxRecordsPerBatch.
Parameters
func – a Python native function that takes an iterator of pandas.DataFrames, and outputs an iterator of pandas.DataFrames.
schema – the return type of the func in PySpark. The value can be either a pyspark.sql.types.DataType object or a DDL-formatted type string.
Maps an iterator of batches in the current DataFrame using a Python native function that takes and outputs a pandas DataFrame, and returns the result as a DataFrame.
The function should take an iterator of pandas.DataFrames and return another iterator of pandas.DataFrames. All columns are passed together as an iterator of pandas.DataFrames to the function and the returned iterator of pandas.DataFrames are combined as a DataFrame. Each pandas.DataFrame size can be controlled by spark.sql.execution.arrow.maxRecordsPerBatch.
Parameters func – a Python native function that takes an iterator of pandas.DataFrames, and outputs an iterator of pandas.DataFrames. schema – the return type of the func in PySpark. The value can be either a pyspark.sql.types.DataType object or a DDL-formatted type string.
‘’’
df = spark.createDataFrame([(1, 21), (2, 30)], ("id", "age"))
def filter_func(iterator):
for pdf in iterator:
yield pdf[pdf.id == 1]
df_mapin = df.mapInPandas(filter_func, schema=df.schema)
print("Input ", "Output")
display_side_by_side(df.toPandas(),df_mapin.toPandas())
Input Output
| id | age |
|---|---|
| 1 | 21 |
| 2 | 30 |
| id | age |
|---|---|
| 1 | 21 |
1h. Co-grouped Map¶
Co-grouped map operations with Pandas instances are supported by DataFrame.groupby().cogroup().applyInPandas() which allows two PySpark DataFrames to be cogrouped by a common key and then a Python function applied to each cogroup.
It consists of the following steps:
Shuffle the data such that the groups of each dataframe which share a key are cogrouped together.
Apply a function to each cogroup. The input of the function is two pandas.DataFrame (with an optional tuple representing the key). The output of the function is a pandas.DataFrame.
Combine the pandas.DataFrames from all groups into a new PySpark DataFrame.
To use groupBy().cogroup().applyInPandas(), the user needs to define the following:
A Python function that defines the computation for each cogroup.
A StructType object or a string that defines the schema of the output PySpark DataFrame.
The column labels of the returned pandas.DataFrame must either match the field names in the defined output schema if specified as strings, or match the field data types by position if not strings, e.g. integer indices. See pandas.DataFrame on how to label columns when constructing a pandas.DataFrame.
Note that all data for a cogroup will be loaded into memory before the function is applied. This can lead to out of memory exceptions, especially if the group sizes are skewed. The configuration for maxRecordsPerBatch is not applied and it is up to the user to ensure that the cogrouped data will fit into the available memory.
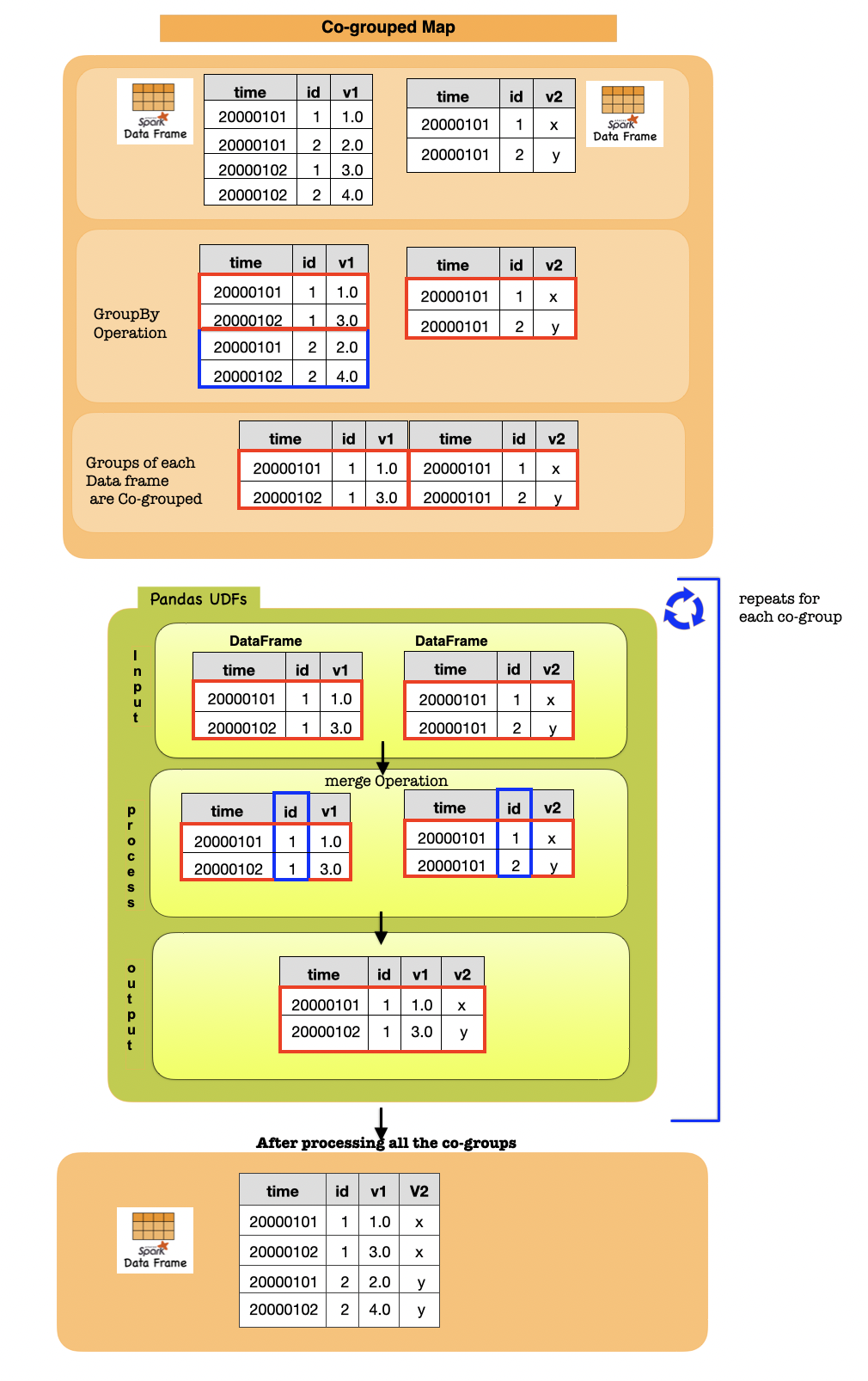
Lets first understand the syntax
Syntax
pyspark.sql.PandasCogroupedOps(gd1, gd2)
Applies a function to each cogroup using pandas and returns the result as a DataFrame.
The function should take two pandas.DataFrames and return another pandas.DataFrame.
For each side of the cogroup, all columns are passed together as a pandas.DataFrame to the user-function and the returned pandas.DataFrame are combined as a DataFrame.
The schema should be a StructType describing the schema of the returned pandas.DataFrame. The column labels of the returned pandas.DataFrame must either match the field names in the defined schema if specified as strings, or match the field data types by position if not strings, e.g. integer indices. The length of the returned pandas.DataFrame can be arbitrary.
Parameters
func – a Python native function that takes two pandas.DataFrames, and outputs a pandas.DataFrame, or that takes one tuple (grouping keys) and two pandas DataFrame
s, and outputs a pandasDataFrame.schema – the return type of the func in PySpark. The value can be either a pyspark.sql.types.DataType object or a DDL-formatted type string. ‘’’
import pandas as pd
df1 = spark.createDataFrame(
[(20000101, 1, 1.0), (20000101, 2, 2.0), (20000102, 1, 3.0), (20000102, 2, 4.0)],
("time", "id", "v1"))
df2 = spark.createDataFrame(
[(20000101, 1, "x"), (20000101, 2, "y")],
("time", "id", "v2"))
def asof_join(l, r):
return pd.merge_asof(l, r, on="time", by="id")
df_out = df1.groupby("id").cogroup(df2.groupby("id")).applyInPandas(
asof_join, schema="time int, id int, v1 double, v2 string")
print(" Input ", "Output")
display_side_by_side(df1.toPandas(),df2.toPandas(), df_out.toPandas())
Input Output
| time | id | v1 |
|---|---|---|
| 20000101 | 1 | 1.0 |
| 20000101 | 2 | 2.0 |
| 20000102 | 1 | 3.0 |
| 20000102 | 2 | 4.0 |
| time | id | v2 |
|---|---|---|
| 20000101 | 1 | x |
| 20000101 | 2 | y |
| time | id | v1 | v2 |
|---|---|---|---|
| 20000101 | 1 | 1.0 | x |
| 20000102 | 1 | 3.0 | x |
| 20000101 | 2 | 2.0 | y |
| 20000102 | 2 | 4.0 | y |
